PBequi unusable with Jackknife at low data precision
An attempt is made to understand why previous experiments with PBequi regression combined with confidence intervals calculated with Jackknife have failed and why, as reported in the previous post of Feb. 6, there are too many rejections of the null hypothesis when the slope is exactly 1.
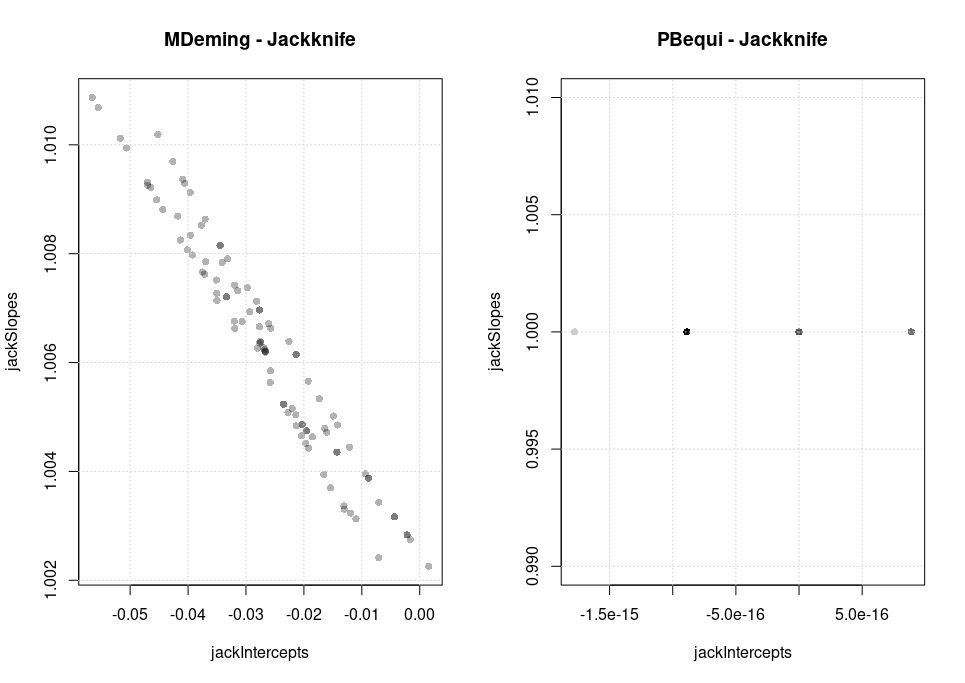
A plot of the individual intercept/slope pairs was plotted in analogy to what is usually done with bootstrap experiments with a data set analysed with PBequi and, as comparison, the MDeming regression of the mcrPioda package (install_github(“piodag/mcrPioda”).
With 2-significant-digit precision of the data one can see how the jackknife variance of slope and intercept for the PBequi regression collapses to zero. On the x-axis we see artefacts caused by the binary representation of the data, on the y-axis not even that and the slopes are all strictly equal to 1. The colour saturation is adjusted to show the overlap of the points.
Thus PBequi can not be paired with Jackknife reliably.
Further the very same data set under bootstrap conditions (BCa as CI choice, but it’s not that relevant here) gives the following picture.
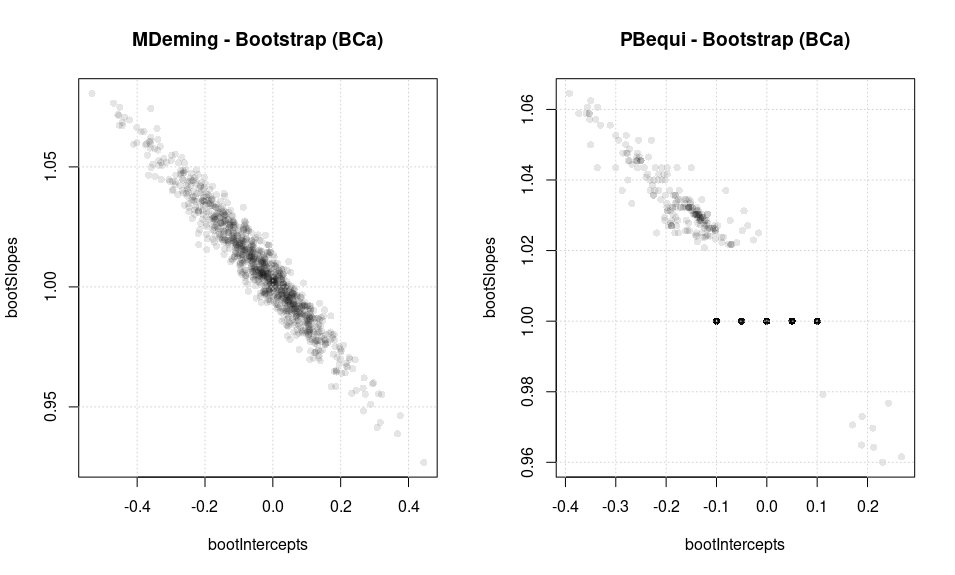
The bootstrap pairs cloud shows high accumulation. A great majority of pairs has slope=1. Moreover, the cloud has a strongly asymmetric aspect. Apparently the slopes > 1 are more likely to occur. Thus PBequi seems to have a bad behaviour also if paired with bootstrap if the data is rounded to low 2x2 digit precision.
The data set was created with set.seed(202402), so the experiment is easily replicable.
The data set generator script is the same reported in the appendix 6.1.4 of the 2021 article, pag. 48.
